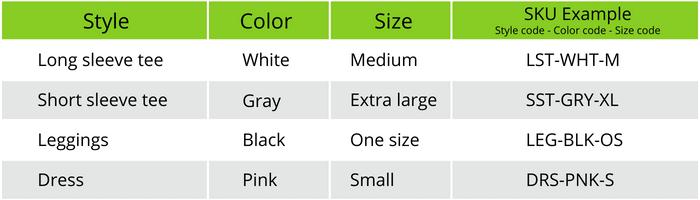
Technical spotlight: Detecting small- and medium-length copy number variants by whole-genome sequencing
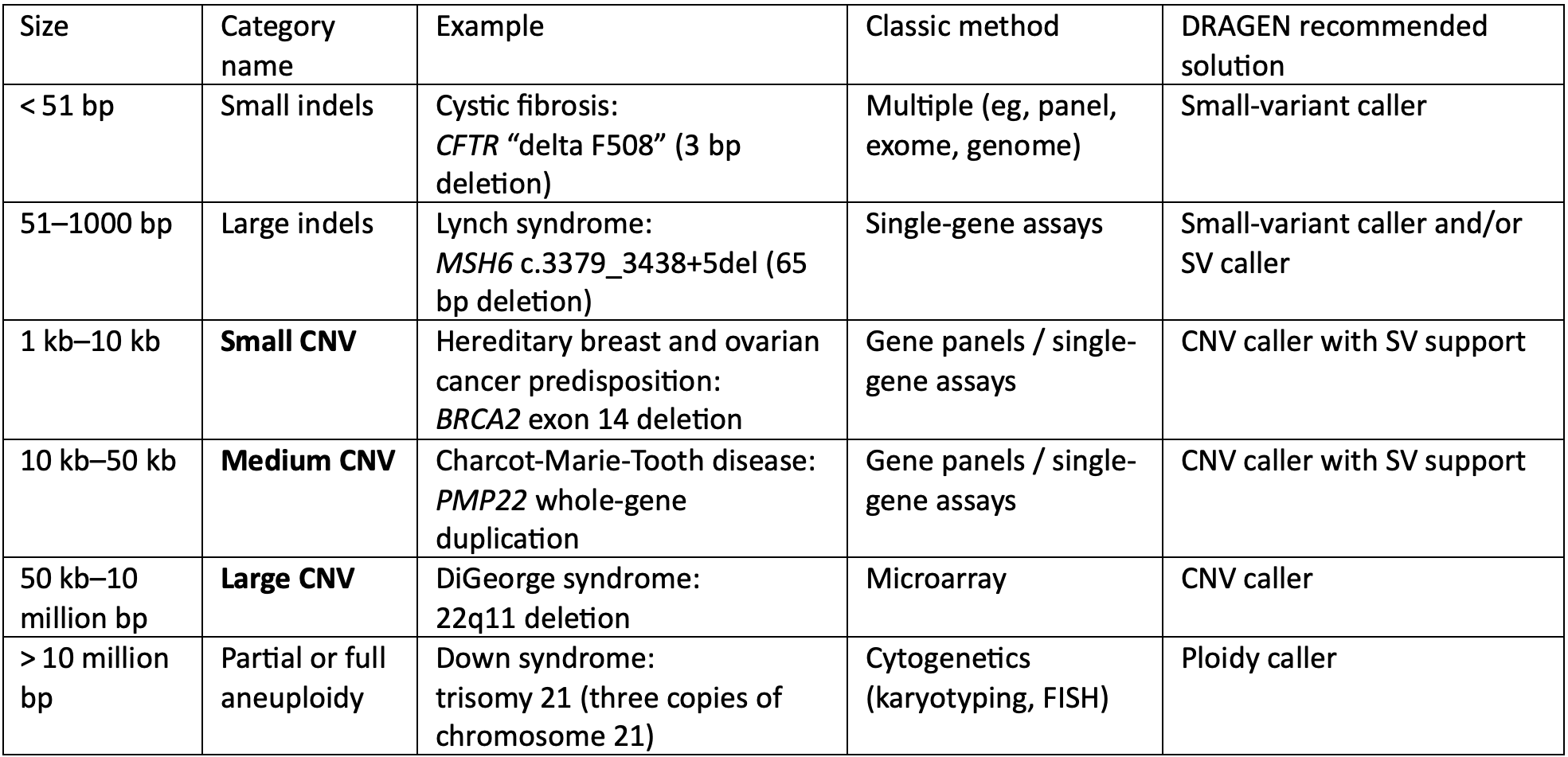
Historically, detecting different sizes of genetic variants has required using multiple different tests. By combining Illumina WGS with secondary analysis algorithms built into the DRAGEN Bio-IT Platform, researchers can achieve high-sensitivity detection of all these different variant types using a mixture of methods described here.

The G Word Genomics England

Genomics Articles Recent genomics discoveries by Illumina scientists

Copy Number Variant Detection Using Next-Generation Sequencing - ScienceDirect

Genomics Research Illumina research & innovation

Rami Mehio on LinkedIn: Edico Genome's team that built DRAGEN in
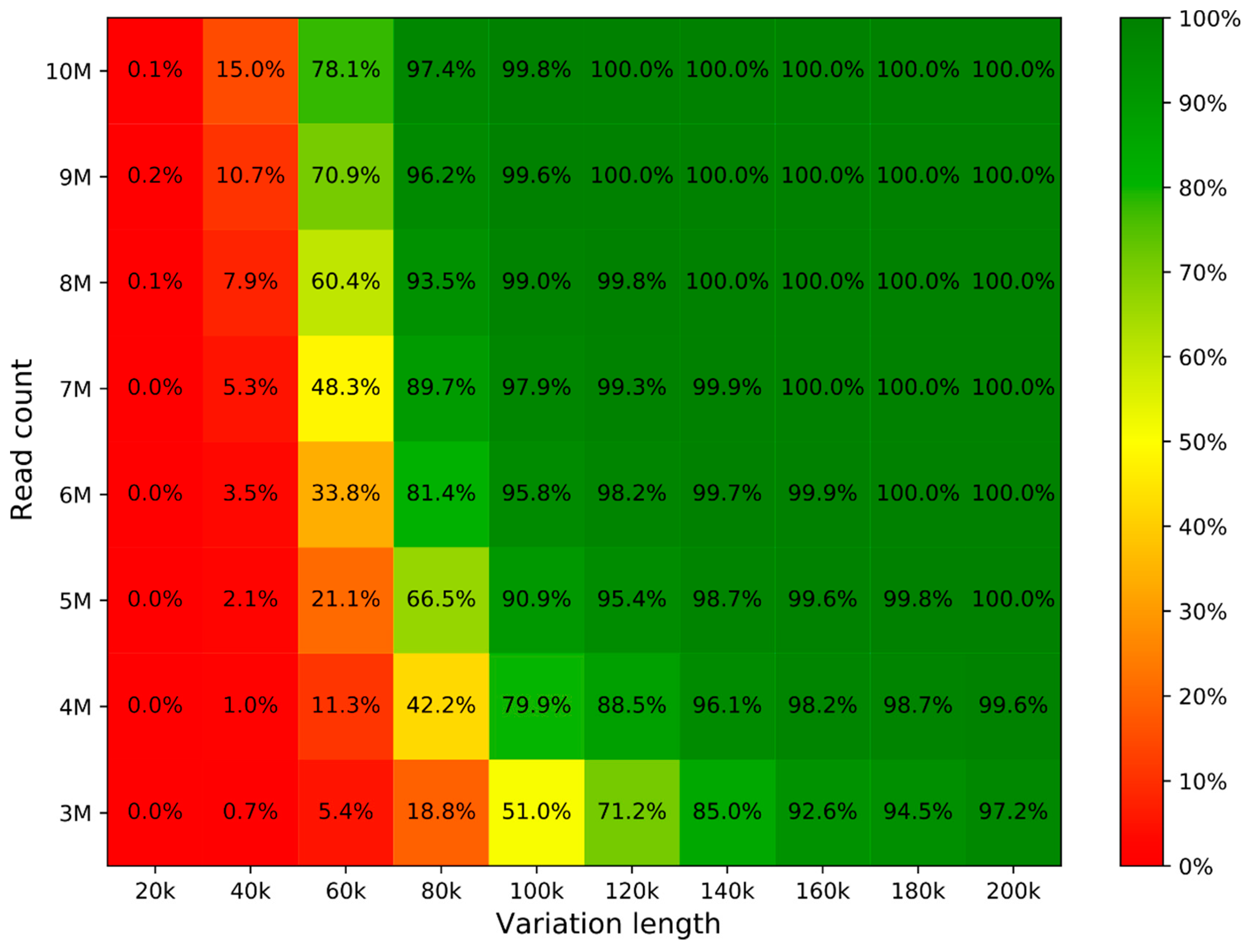
Diagnostics, Free Full-Text
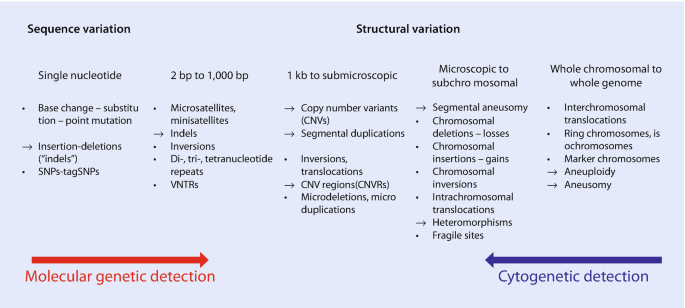
SNPs, GWAS, CNVs: Informatics for Human Genome Variations

Rami Mehio on LinkedIn: Using whole-genome sequencing to evaluate

Rosy Volpi on LinkedIn: Session 2: Expanding Frontiers of Genomic
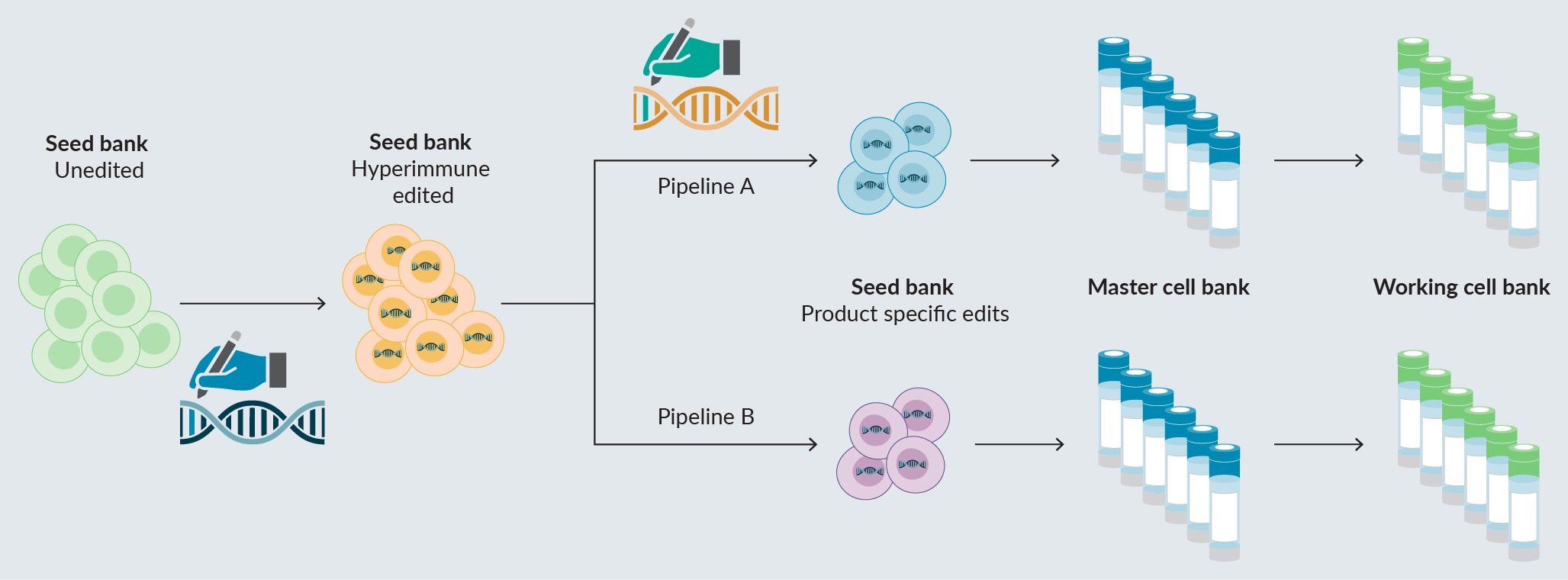
Considerations for development of gene-edited PSC-based therapies

Rami Mehio on LinkedIn: Video: Targeting the right drug, at the

Maria Martínez-Fresno Moreno on LinkedIn: Comprehensive and

SV detection strategies using short reads. (A) Read-depth-based method

Copy Number Variation (CNV) Analysis